Big data is big business in the life sciences, attracting lots of money and prestige. It's also relatively young; the move toward big data can be traced back to 1990, when researchers joined together to sequence all three billion letters in the human genome. That project was completed in 2003 and since then, the life sciences have become a data juggernaut, propelled forward by sequencing and imaging technologies that accumulate data at astonishing speeds.
The National Ecological Observatory Network, funded by Congress with $434m, will equip 106 sites in the United States with sensors to gather ecological data all day, every day, for 30 years when it starts operating in three years. The Human Brain Project, supported by $1.6 billion from the European Union, intends to create a supercomputer simulation of a working human brain, including all 86 billion neurons and 100 trillion synapses. The International Cancer Genome Consortium, 74 research teams across 17 countries spending an estimated $1 billion, is compiling 25,000 tumour genome sequences from 50 types of cancers.
But not all scientists think bigger is better. More than 450 researchers have already signed a public letter criticising the Human Brain Project, citing a "significant risk" that the project will fail to meet its goal. One neuroscientist called the project "a waste of money", while another bluntly said the idea of simulating the human brain is downright "crazy". Other big data projects have also been criticised, especially for cost and lack of results.
The main concern of the critics is that expensive, massive data sets on biological phenomenon – including the brain, the genome, the biosphere, and more – won't necessarily lead to scientific discoveries. "One of the problems with ideas about big data is the implicit notion that simply having lots of data will answer questions for you, and it won't," says J Anthony Movshon, a neuroscientist at New York University. Large data sets are only useful when combined with the right tools and theories to interpret them, he says, and those have largely been lacking in the life sciences.
That's one reason biological data is piling up far faster than it is being analysed. "We have an inability to slow down and focus," says Kenneth Weiss, an evolutionary geneticist at Penn State University. "I wouldn't say big data is bad, but it's a fad, and we're not learning a lesson from it."
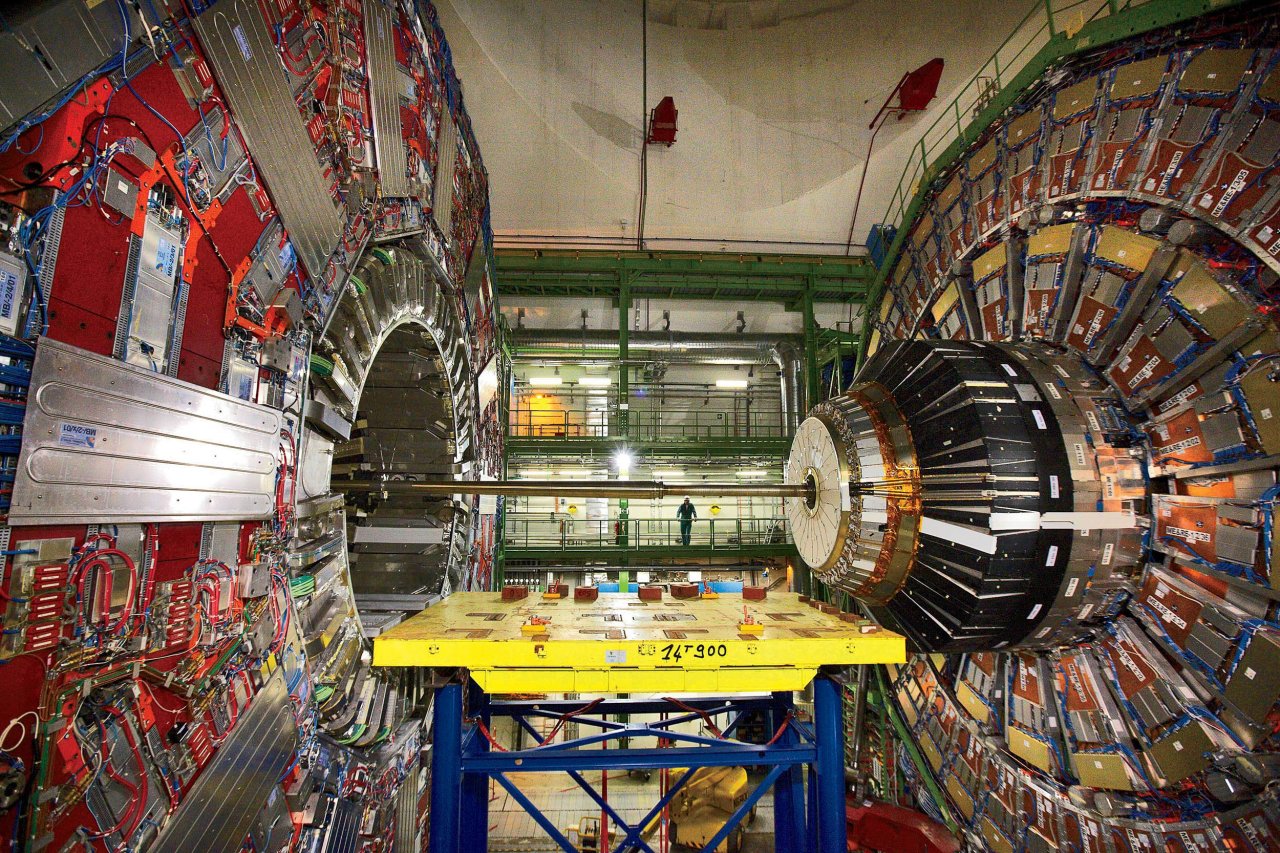
Other areas of science, such as physics and astronomy, also have a rich history of big data, as well as the organisation and infrastructure to use that data. Take the Hubble Space Telescope, which has made one million observations, amounting to more than 100 terabytes of data, since its launch in 1990. More than 10,000 scientific articles have been published using that data, including the discovery of dark energy and the age of the universe. Or consider the Large Hadron Collider, a particle accelerator that produces tens of terabytes of data each night. In 2012, that data confirmed the existence of the Higgs boson, also called the "God particle", among other high-profile discoveries in particle physics.
In the life sciences, on the other hand, data collection seems to have gone ahead without the ability to determine which types of data are most useful, how to share it, or how to reproduce results. Research and funding institutions recognise this limitation, says Philip Bourne, associate director for data science at the National Institutes of Health, and they are working to set aside funding and manpower to find ways to make data usable. Bourne is optimistic: "Making full use of very large amounts of data takes time, but I think it will come," he says.
Bourne and others, like David Van Essen, lead investigator of the $40m NIH-funded Human Connectome Project (HCP), believe that gathering data first and asking questions second is an exciting way to make discoveries about the natural world. The HCP, a consortium of 36 investigators at 11 institutions, is a big data effort to map the connections in the brain using high-resolution brain scans and behavioral information from 1,200 adults. According to the project's website, the HCP data set will "reveal much about what makes us uniquely human and what makes every person different from all others." On the other hand, there's not a single hypothesis in sight.
This is a fundamentally different way of doing science from hypothesis-driven experiments, the traditional bedrock of the scientific method, and many researchers have their doubts about it.
"Science depends upon predictions being generated and those hypotheses being tested," says ecologist Robert Paine of the University of Washington. "Mega models won't bring us to the promised land." Others say the effort required to gather the data simply doesn't warrant the price tag: "The idea that you should collect a lot of information because somewhere in this chaff is a little bit of wheat is a poor case for using a lot of money," says Movshon.
Movshon may be justified in his criticism: so far, novel discoveries from massive data sets have been few and far between, admits Bourne. In some cases, the results have been downright disappointing: large analyses of the genomes of thousands to hundreds of thousands of individuals to identify genes associated with disease have become very popular – publications of these so-called "genome-wide association studies" have increased from fewer than 10 in 2005 to more than 1,750 today – but are often unsuccessful. They have identified few meaningful genes associated with disease.
But there are still discoveries to come, if researchers connect the right data with the right models, argues Drew Purves, head of computational ecology at Microsoft Research. Purves and his team recently unveiled the first virtual model of the Earth's ecosystem, a silicon representation of all life on the planet. They now plan to stock the Madingley Model (named after the British village where the idea was hatched) with as much data as they can get their hands on, from species interactions to climate data and more. While other ecologists have criticised the project as a waste of resources, Purves laughs at the claim. Three postdoctoral researchers spent three years developing the model, he says. "We're the tip of the iceberg. Are we seriously saying the entire global scientific field of ecology can't spare three scientists for three years to aggregate it all together into a picture of the whole?"
Critics also raise concerns about whether discoveries from such data sets can be reproduced. For example, say the International Cancer Genome Consortium uses its 25,000 tumour genomes to discover that one gene causes 99% of all cancers (it doesn't). There is no second database of comparable tumour sequences to verify that finding. "If [a team] finds something novel, how are they going to test its reality and distinguish it from simply being an artefact of the complexity of their database or model?" asks Paine. Van Essen contends that researchers can take steps to show the findings are reproducible, such as repeating small parts of a large experiment to see if the results match the original data, or dividing a data set in half and doing the same experiment on each half to see if the results agree.
Finally, perhaps the biggest worry about big data is money. With a blank check, scientists would be happy to collect any and all data. But research funding has been tight since 2009, and there is growing concern that money and manpower spent on unproven big data projects is money and manpower taken from smaller, hypothesis-driven experiments with a history of success.
"There's a lot of honest concern by many neuroscientists [that] if the pendulum were to swing too far, that individual investigator-inspired projects might become a threatened species," says Van Essen. "I personally don't think that will happen." Neither does Bourne at the NIH, who must decide what proportion of government research funding should be directed toward big data initiatives. The NIH is conscious of the concern and will work to balance funding between the two, he says.
"Biological research is becoming a data enterprise," says Bourne. "It's a new way of doing things, and new things are always greeted, perhaps rightly so, with skepticism. Some rise and some don't." Bourne, however, is a believer in big data. "There's no doubt in my mind this will rise," he says.
Meanwhile, sceptics wait to see if big data will live up to the hype – and be worth the cost. "We all agree that moving forward [in science] is important, though we will always argue about the details of which is the best approach," says Movshon. "You can call me a Luddite . . . . but there are good reasons to be concerned."